Here are software we have developed or contributed to
PhotoFiTT (2024)
PhotoFiTT (Phototoxicity Fitness Time Trial) is an integrated framework combining a standardised experimental protocol with advanced image analysis to quantify light-induced cellular stress in label-free settings. PhotoFiTT leverages machine learning and cell cycle dynamics to analyse mitotic timing, cell size changes, and overall cellular activity in response to controlled light exposure.


CellTracksColab (2024)
CellTracksColab is an online platform tailored to simplify the exploration and analysis of tracking data by leveraging the free, cloud-based computational resources of Google Colab. CellTracksColab facilitates the amalgamation and analysis of results across multiple fields of view, conditions, and repeats, ensuring a holistic dataset overview. CellTracksColab also harnesses the power of high-dimensional data reduction and clustering, enabling researchers to identify distinct behavioral patterns and trends without bias. Finally, CellTracksColab also includes specialized analysis modules enabling spatial analyses (clustering, proximity to specific regions of interest).
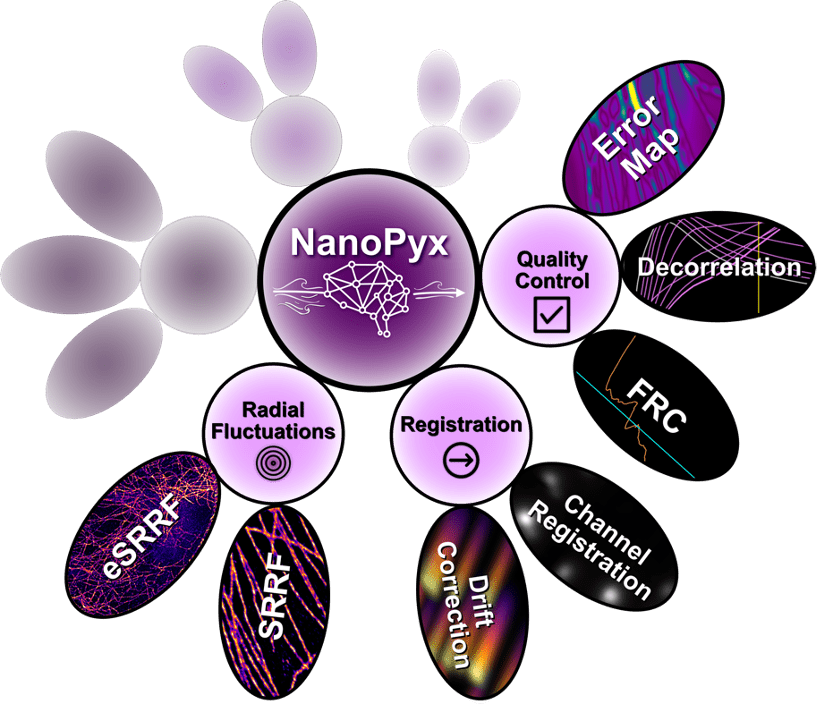
NanoPyx (2024)
NanoPyx is a library specialized in the analysis of light microscopy and super-resolution data. It is a successor to NanoJ, which is a Java library for the analysis of super-resolution microscopy data. NanoPyx focuses on performance, by heavily exploiting cython aided multiprocessing and simplicity. It implements methods for the bioimage analysis field, with a special emphasis on those developed by the Henriques Laboratory. It will be distributed as a Python Library and also as Codeless Jupyter Notebooks, that can be run locally or on Google Colab, and as a napari plugin.

DL4MicEverywhere (2024)
DL4MicEverywhere is a platform that offers researchers an easy-to-use gateway to cutting-edge deep learning techniques for bioimage analysis. It features interactive Jupyter notebooks with user-friendly graphical interfaces that require no coding skills. The platform utilizes Docker containers to ensure portability and reproducibility, guaranteeing smooth operation across various computing environments.
DL4MicEverywhere extends the capabilities of ZeroCostDL4Mic by allowing the execution of notebooks either locally on personal devices like laptops or remotely on diverse computing platforms, including workstations, high-performance computing (HPC), and cloud-based systems. It currently incorporates numerous pre-existing ZeroCostDL4Mic notebooks for tasks such as segmentation, reconstruction, and image translation.
eSRRF (2023)
eSRRF provides an accessible super-resolution approach, maximizing information extraction across varied experimental conditions while minimizing artifacts. Its optimal parameter prediction strategy is generalizable, moving toward unbiased and optimized analyses in super-resolution microscopy.
Fast4DReg (2023)
Fast4DReg is a Fiji plugin for 2D and 3D video and can correct drift in all x-, y-, and z-directions. Fast4DReg creates intensity projections along both axes, estimates their drift using cross-correlation-based drift correction, and then translates the video frame by frame. Additionally, Fast4DReg can be used for aligning multi-channel 2D or 3D images, which is particularly useful for instruments that suffer from a misalignment of channels.
TrackMate (2022)
TrackMate is your buddy for your everyday tracking.
TrackMate is an automated tracking software that analyzes bioimages and is distributed as a Fiji plugin.
TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Read our article here.
Automated cell tracking using StarDist and TrackMate. Read our paper here.
ZeroCostDL4Mic (2021)
ZeroCostDL4Mic allows the use of popular Deep Learning neural networks capable of carrying out tasks such as image segmentation and object detection (using U-Net, StarDist, and YOLOv2), image denoising and restoration (using CARE and, Noise2Void), super-resolution microscopy (using Deep-STORM) and image-to-image translations (using Label-free prediction fnet, pix2pix, and CycleGAN).
Read our paper and access the ZeroCostDL4Mic platform.
SRRF TFM (2020)
we propose a simplified protocol and imaging strategy that enhances the output of traction force microscopy by increasing i) achievable bead density and ii) the accuracy of bead tracking. Our approach relies on super-resolution microscopy, enabled by fluorescence fluctuation analysis. Our pipeline can be used on spinning-disk confocal or widefield microscopes and is compatible with available analysis software.
Read our paper.
FiloMap (2019)
ImageJ and R scripts used to map the localization of proteins within filopodia and generate filopodia maps.
Read our paper here.
Step-by-step protocols on how to use FiloMap to compare the localization of multiple proteins within filopodia.
Access our article here.
Find FiloMap on GitHub.
FiloQuant (2017)
The filopodia quantification plugin
FiloQuant is a user-friendly tool for the automated detection and quantification of filopodia properties such as length and density. We developed FiloQuant as a plugin for Fiji. Read our paper.
Step-by-step protocols on how to use FiloQuant to quantify filopodia
and filopodia-like protrusions from microscopy images. Access our article here.
Find FiloQuant on GitHub.
You must be logged in to post a comment.